A new framework for precise in vivo functional analysis of DNA variants
Press release by Irene Faipò and authors (Alex Cornean, Jakob Gierten, Bettina Welz, Juan Luis Mateo, Thomas Thumberger, Joachim Wittbrodt)
A highly interdisciplinary research group led by the Principal Investigator of the Cluster of Excellence “3D Matter Made to Order” (3DMM2O), Prof. Dr. Joachim Wittbrodt, developed a state-of-the-art framework for in vivo base editing. This framework enables the precise functional analysis of DNA variants. Geneticist and data scientist Alex Cornean, pediatric cardiologist Dr. Jakob Gierten, and biologist Bettina Welz collaborated closely with computer scientist Prof. Dr. Juan Luis Mateo, and developmental biologist Dr. Thomas Thumberger. They synergised to simplify how to answer questions on the functions of a particular genetic component in a model organism, by performing specific changes to this component.
Single nucleotide variants (SNVs) are small but abundant changes of the DNA that contribute to the traits of any individual. However, they can also increase the susceptibility to diseases, including ocular and cardiovascular ones. Therefore, understanding the effects of these DNA variants is key to improve personalized patients’ clinical care. Model organisms, such as the Japanese rice fish medaka, and zebrafish, are ideal to examine the significance of a variant because their organs can fully develop in just a few days. Besides, the transparent embryos are easy to observe under a microscope, and relevant areas can be highlighted using fluorescent proteins.
The introduction of DNA variants into an organism by base editing has been restricted by limited in vivo efficiencies of base editors and a lack of bioinformatic tools to design these DNA changes. To overcome these bottlenecks, an interdisciplinary group of scientists from Heidelberg University and the University of Oviedo (Spain), developed a cutting-edge framework for base editing. This framework allows to change the DNA building blocks adenine (ABE) and cytosine (CBE) in medaka and zebrafish, or in virtually any other model organism. The researchers identified suitable, state-of-the-art base editors with remarkable in vivo efficiencies that enabled them to assess phenotypes rapidly and exhaustively in fish embryos. Seeking to simplify this process, the team developed the new open-source, online base editing tool ACEofBASEs (a careful evaluation of base edits). ACEofBASEs accelerates decision-making because it allows researchers to streamline sgRNA design and perform an off-target evaluation. Together, these efforts converge into an ideal framework for scalable validation studies.
The team applied this framework in medaka and zebrafish to edit several test genes with well documented functions. Initially, they computationally designed single base changes for eye pigmentation or transgenic GFP genes. These variations would result in single amino acid mutations or a protein truncation which abolishes the gene’s function. Later, the team introduced single base mutations into the target genes in vivo and demonstrated the expected functional gene inactivation using light microscopy, i.e., the loss of eye pigmentation or inactivation of transgenic GFP signals (Figure 1). Besides, base editing in the genes encoding troponin T and the potassium channel ERG faithfully recreated expected cardiac phenotypes. The group showed that base editing allows one to zoom into specific protein domains and probe the functional relevance of single amino acid changes. In the case of the cardiac potassium channel ERG, a single mutation stopped the beating of the heart ventricle and drastically changed its morphology.
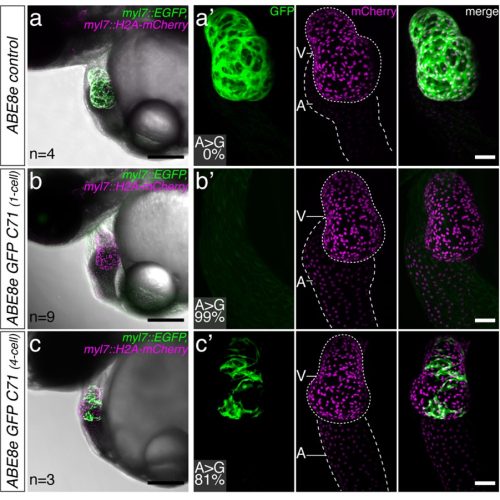
This approach enabled the researchers to validate single missense mutations initially associated with human congenital heart defects in medaka. The gene mutations that were introduced into model organisms showed visible effects on the morphology and functions of embryonic hearts. These results highlight the potential of targeted base editing for disease modelling: “Our achievement was to translate an association into causality”, emphasised Alex Cornean, Jakob Gierten, and Bettina Welz.
This interdisciplinary research group closed the gap between SNV discovery and validation and paved the way for detailed mechanistic downstream studies in multiple additional applications, ranging from immunology and glycosylation to stem cells and development.
Further information:
- Cluster of Excellence "3D Matter Made to Order” (3DMM2O)
- ACEofBASEs
- CRISPR-Cas target online predictor
- Precise in vivo functional analysis of DNA variants with base editing using ACEofBASEs target prediction Alex Cornean, Jakob Gierten, Bettina Welz, Juan Luis Mateo, Thomas Thumberger, Joachim Wittbrodt. eLife 2022; 11:e72124 DOI: 10.7554/eLife.72124




