Boosting the efficiency of gene editors with “hei-tag”
Press release by Irene Faipò and authors (Thomas Thumberger, Tinatini Tavhelidse-Suck, Jose Arturo Gutierrez-Triana, Alex Cornean, Rebekka Medert, Bettina Welz, Marc Freichel, Joachim Wittbrodt)
Along the lines of the strong interdisciplinary approach of the Cluster of Excellence “3D Matter Made to Order” (3DMM2O), a research group led by cluster’s spokesperson Prof. Dr Jochen Wittbrodt investigated how to change the DNA more efficiently to perform genetic alterations of certain loci. An interdisciplinary team comprising developmental biologists Dr. Thomas Thumberger and Dr. Tinatini Tavhelidse-Suck from the Centre for Organismal Studies, and molecular biologist Dr. Jose Arturo Gutierrez-Triana (currently, Universidad Industrial de Santander, Colombia) combined efforts with Dr. Rebekka Medert and Prof. Dr Marc Freichel from the Institute of Pharmacology of Heidelberg University. They developed a new molecular tag as a straightforward addition to already existing genome editing tools. The new “high efficiency-tag” (hei-tag) boosts both targeting efficiency in cell cultures grown in the lab and genome editing in entire developing organisms.
The genetic code stored within DNA provides cells with the instructions they need to carry out their role in the body. Since any changes to these genes can completely alter how a cell behaves, scientists have developed different tools that enable them to modify the genome of cells in culture or of entire living organisms. This includes both the Cas9 enzyme, which can cut DNA at specific sites, as well as multiple base editors allowing researchers to perform changes on bits of genetic code without cutting DNA. While a wide range of Cas9 enzymes and base editors is currently available, identifying a tool that can effectively modify an organism’s genome at the right time of development can be rather challenging.
One needs to keep in mind that all the cells in an organism arise from a single fertilised cell. Consequently, if this cell is genetically edited, all its subsequent daughter cells will contain that same genetic modification. Even though genome editing tools such as CRISPR/Cas9 are key for basic research and translational approaches, most of these tools only work efficiently at later stages of development, thus creating a mosaic organism consisting of both edited and non-edited cells. To tackle this aspect, the team identified a new “high efficiency-tag” (hei-tag) which boosts the activity of CRISPR/Cas genome editing tools already when supplied as mRNA. The “hei-tag” enhances the targeting efficiency of Cas9 enzymes, possibly by promoting their nuclear localization. This results in an even distribution of edited genes spread throughout the whole organism (Figure 1). “A more homogenous editing not only tackles mosaicisms in developing organisms, but also makes the phenotype more reliable”, highlight Thomas Thumberger, Tinatini Tavhelidse-Suck, and Jose Arturo Gutierrez-Triana.
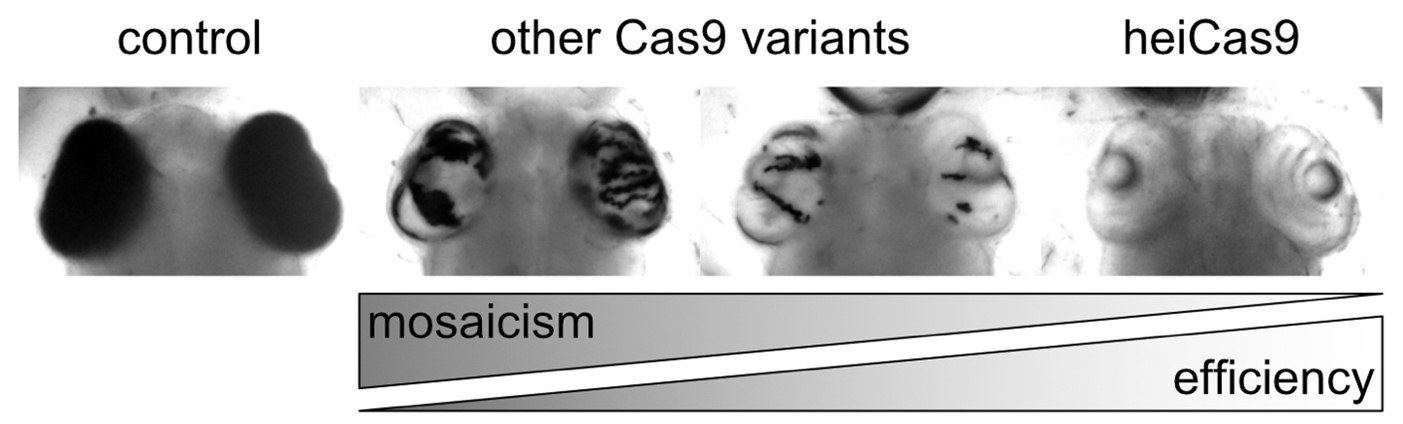
For developmental biologists, it is critical to be able to edit a gene as early in development as possible, in order to understand how that gene will affect the organism. By simply adding the “hei-tag” to already available tools, scientists can upgrade existing and highly adapted systems. This has high potential to pave the way for advances in medical research aiming at replacing faulty genes with fully functioning ones.
Further information:
- Cluster of Excellence "3D Matter Made to Order” (3DMM2O)
- ACEofBASEs
- CRISPR-Cas target online predictor
- Boosting targeted genome editing using the hei-tag, Thomas Thumberger, Tinatini Tavhelidse-Suck, Jose Arturo Gutierrez-Triana, Alex Cornean, Rebekka Medert, Bettina Welz, Marc Freichel, Joachim Wittbrodt, eLife 2022;11:e70558 DOI: 10.7554/eLife.70558
